13.4 Approaches for Vaccine Development Against SARS‐Cov‐2
The elucidation of the genome organization and functional domains of S protein for SARS-CoV (Figure 13.4.1), achieved through the work of scientists and geneticists all over the globe, has facilitated a deep understanding of the mode of action of this virus which has led to the development of a myriad of vaccine and drug candidates in an effort to mitigate the spread of this virus, as well as to assist in the diagnosis and management of infected patients.
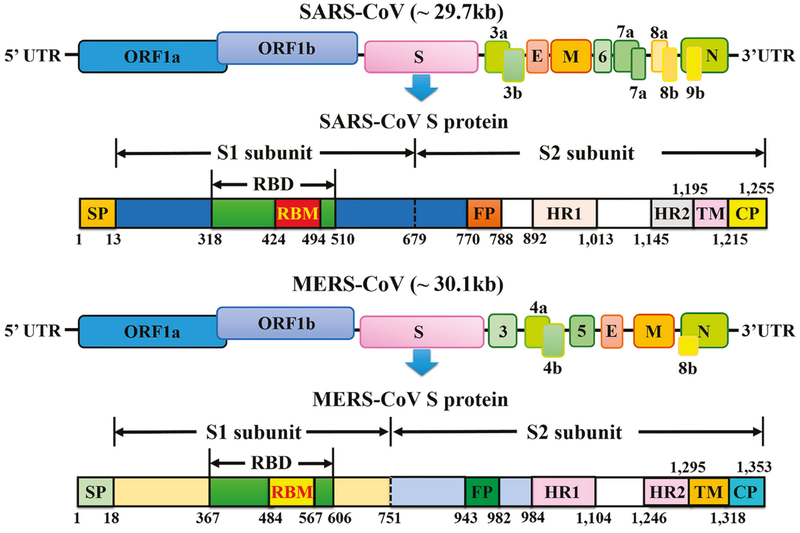
Figure 13.4.1 is a schematic representation of the genome organization and functional domains of S protein for SARS-CoV and MERS-CoV. The single-stranded RNA genomes of SARS-CoV and MERS-CoV encode two large genes, the ORF1a and ORF1b genes, which encode 16 non-structural proteins (nsp1–nsp16) that are highly conserved throughout coronaviruses. The structural genes encode the structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), which are common features to all coronaviruses. The accessory genes (shades of green) are unique to different coronaviruses in terms of number, genomic organization, sequence, and function. The structure of each S protein is shown beneath the genome organization. The S protein mainly contains the S1 and S2 subunits. The residue numbers in each region represent their positions in the S protein of SARS and MERS, respectively. The S1/S2 cleavage sites are highlighted by dotted lines. SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; CP, cytoplasm domain; FP, fusion peptide; HR, heptad repeat; RBD, receptor-binding domain; RBM, receptor-binding motif; SP, signal peptide; TM, transmembrane domain.
Different approaches for the development of vaccine candidates against SARS‐Cov‐2 have, and are being, developed and utilized. Figure 13.4.2 summarizes these methods:
-
- Potential vaccines under development, involve five leading platforms (inactivated virus, protein subunit, DNA, RNA, and non‐replicating viral vector), as depicted.
- Intact SARS‐CoV‐2 is neutralized by treatment with radiation to cease its ability to infect and replicate, while preserving the induction of an immune response.
- A plasmid DNA is genetically engineered with the S, M, and N genes of SARS‐CoV‐2 encoding the respective proteins that may facilitate an immune response.
- A replication‐defective Adenovirus (Ad) vector is genetically engineered to express SARS‐Cov‐2 spike (S) protein.
- An mRNA (replication‐defective) that encodes the S protein of SARS‐CoV‐2 is encapsulated in a lipid nanoparticle (LNP), which, when injected, induces the body cells to produce the spike protein and direct the immune response.
- Spike protein‐encoding (S) gene of SARS‐CoV‐2 was isolated and genetically engineered into a baker’s yeast, producing the spike protein antigens when grown. The produced S antigens can then be collected and purified.
Watch the video below called, There are four types of COVID-19 vaccines: here’s how they work, by Gavi, the Vaccine Alliance (2020) on YouTube, which gives a brief overview of the main types of COVID-19 vaccines and the mechanism by which they bring about immunity in a patient.
Media Attributions
- Figure 13.4.1 SARS-CoV MERS-CoV genome organization and S-protein domains by Song, et al. (2019), CC BY 4.0, via Wikimedia Commons
- Figure 13.4.2 Different approaches for the development of vaccine candidates against SARS‐Cov‐2 by Faheem et al. (2021), CC BY 4.0, via Wikimedia Commons
References
Faheem, M. S. et al. (2021). Platforms exploited for SARS-CoV-2 vaccine development. Vaccines 9(1), 11. https://doi.org/10.3390/vaccines9010011
Gavi, the Vaccine Alliance. (2020, December 18). There are four types of COVID-19 vaccines: here’s how they work (video file). YouTube. https://www.youtube.com/watch?v=lFjIVIIcCvc
Song Z et al. (2019, January 14). From SARS to MERS, Thrusting coronaviruses into the spotlight. Viruses 2019, 11(1), 59. https://doi.org/10.3390/v11010059
Long Descriptions
- Figure 13.4.1 Schematic representation of the genome organization and functional domains of S protein for SARS-CoV and MERS-CoV. The single-stranded RNA genomes of SARS-CoV and MERS-CoV encode two large genes, the ORF1a and ORF1b genes, which encode 16 non-structural proteins (nsp1–nsp16) that are highly conserved throughout coronaviruses. The structural genes encode the structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), which are common features to all coronaviruses. [Back to Figure 13.4.1]
- Figure 13.4.2 Different Approaches for the Development of Vaccine Candidates Against SARS‐Cov‐2. (A) Potential vaccines under development involve five leading platforms (inactivated virus, protein subunit, DNA, RNA, and non‐replicating viral vector), as depicted. (B) Intact SARS‐CoV‐2 is neutralized by treatment with radiation to cease its ability to infect and replicate, while preserving the induction of an immune response. (C) A plasmid DNA is genetically engineered with the S, M, and N genes of SARS‐CoV‐2 encoding the respective proteins that may facilitate an immune response. (D) A replication‐defective Adenovirus (Ad) vector is genetically engineered to express SARS‐Cov‐2 spike (S) protein. (E) An mRNA (replication‐defective) that encodes the S protein of SARS‐CoV‐2 is encapsulated in a lipid nanoparticle (LNP), which, when injected, induces the body cells to produce the spike protein and direct the immune response. (F) Spike protein‐encoding (S) gene of SARS‐CoV‐2 was isolated and genetically engineered into a baker’s yeast, producing the spike protein antigens when grown. The produced S antigens can then be collected and purified. [Back to Figure 13.4.2]

