2.1 Introduction
Learning Objectives
- Describe Mendel’s Principle of Independent Assortment.
- Analyze parental and offspring phenotypes to determine patterns of inheritance for two traits.
- Apply probability and branch diagrams to determine genotypic and phenotypic ratios in dihybrid and trihybrid crosses.
- Apply a test cross to identify genotypes in individuals expressing two dominant traits.
- Apply proper use of modern genetic terminology.
- Outline the concept of Linkage (resulting in distortion of the ratios expected from independent assortment).
Having determined from monohybrid crosses that genes are inherited according to the Law of Segregation, Mendel looked at the simultaneous inheritance of two or more unrelated traits. He considered how two pairs of alleles would segregate into a dihybrid individual (i.e., a plant that is heterozygous for two genes). Mendel’s Law of Independent Assortment states that during gamete formation, alleles at separate loci segregate independently, and this produces characteristic genotypic and phenotypic ratios. As such, the principles of genetic analysis that we have described for a single locus in Chapter 1 will be extended to the study of alleles at two loci in this chapter. The analysis of two loci in the same cross provides information for genetic mapping and testing gene interactions.
Take a look at the following YouTube video, Law of Independent Assortment, by AK Lecture Series (2015) on YouTube.
Before Mendel’s 1865 publication, blended inheritance was the accepted model to explain the transmission of traits. It was Mendel’s work that established that heritable traits were controlled by discrete factors, which we now call alleles, in a particulate inheritance model. At the time, it was an important question as to whether heritable traits, controlled by discrete factors, were inherited independently of each other? To answer this, Mendel took two apparently unrelated traits, such as seed shape and seed colour, and studied their inheritance together in one individual. For example, he studied two variants of each trait: seed colour was either green or yellow, and seed shape was either round or wrinkled. (He studied seven traits in all, each on a different chromosome.) When either of these traits was studied individually, the phenotypes segregated in the classical 3:1 ratio among the progeny of a monohybrid cross (Figure 2.1.1), with ¾ of the seeds green and ¼ yellow in one cross, and ¾ round and ¼ wrinkled in the other cross. Would this be true when both hybrids were in the same individual?

Like in the previous Chapter 1, we will first walk through how a dihybrid cross works on at the DNA level, and then we will explain the results that Mendel saw that led him to his law, the Law of Independent Assortment.
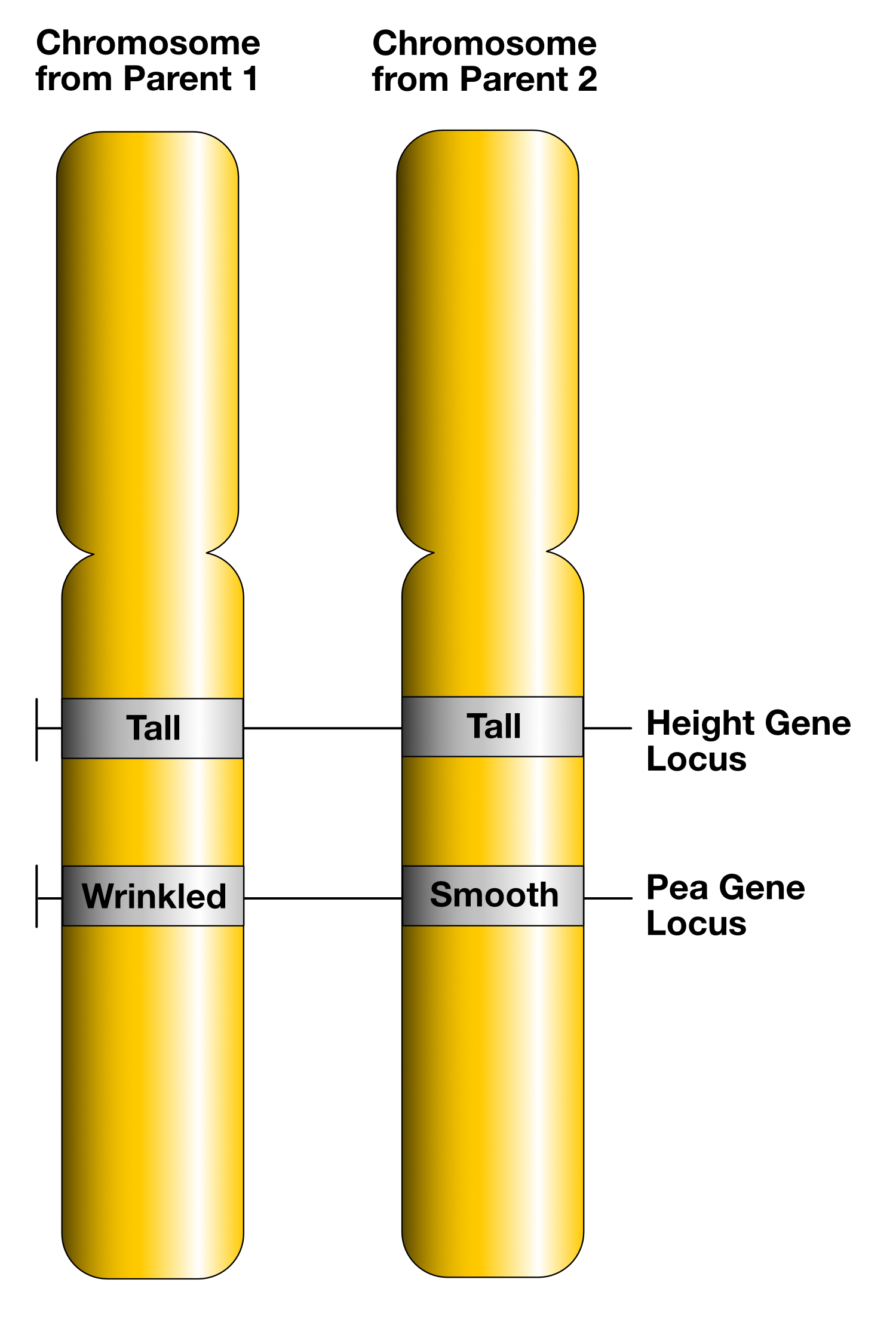
When dealing with alleles at two different loci, we have to use nomenclature that makes the arrangement clear. There are three possible arrangements: Both loci are on the same chromosome (AB/ab), different chromosomes (A/a; B/b) as shown for example in Figure 2.2, or unknown (AaBb).
Media Attributions
- Figure 2.1.1 Original by Deyholos (2017), CC BY-NC 3.0, via Biology Libretexts
- Figure 2.1.2 Gene Loci and Alleles by Keith Chan, CC BY-SA 4.0, via Wikimedia Commons
References
AK Lectures. (2015, January 2). Law of independent assortment [Video file]. YouTube. https://www.youtube.com/watch?v=VjPjyLwcYYQ
Deyholos, M. (2017). Figure 2. Monohybrid crosses involving two distinct traits in peas [digital image]. In Locke, J., Harrington, M., Canham, L. and Min Ku Kang (Eds.), Open Genetics Lectures, Fall 2017 (Chapter 17, p. 1). Dataverse/ BCcampus. http://solr.bccampus.ca:8001/bcc/file/7a7b00f9-fb56-4c49-81a9-cfa3ad80e6d8/1/OpenGeneticsLectures_Fall2017.pdf
Keith Chan. (2015, July 23). Gene Loci and Alleles [digital image]. Wikimedia Commons. https://commons.wikimedia.org/wiki/File:Gene_Loci_and_Alleles.png
Long Descriptions
- Figure 2.1.1 Two sets of genetic crosses, using green- and yellow-colored seeds. The first genetic cross shows the production of F1 and F2 generations from parental pure lines. In this cross, one pure breeding round seeded parent is crossed with one pure breeding wrinkled seeded parent. The F1 generation is produced, and all have round seeds and are heterozygous for this trait. The subsequent F2 generation produces round and wrinkled seeds in the typical 3:1 monohybrid ratio. The other genetic cross shows the same type of cross, except this time for yellow and green seeds, with yellow as the dominant trait, and green as the recessive trait. [Back to Figure 2.1.1]
- Figure 2.1.2 Two homologous chromosomes colored in yellow with two gene loci shown on these chromosomes — one for plant height and the other for pea shape. Both chromosomes contain the allele for tall height, but the first has the allele for wrinkled seeds, and the other has the allele for round seeds. [Back to Figure 2.1.2]

