12.1 Introduction
Learning Objectives
- Distinguish among genetic (recombination), cytogenetic (metaphase chromosome), and physical maps.
- Differentiate chromosomes cytologically based on their length, centromere position, and banding patterns when stained with dyes.
- Discuss the ultimate physical map which is the DNA sequence of the whole chromosome or genome.
In this chapter, we will look at the larger picture of chromosomes and whole genomes, and the various methods used to visualize them. Many types of mapping techniques are available to identify single genes responsible for disorders, such as ankylosing spondylitis and cystic fibrosis, as well as the multiple genes responsible for common conditions, such as cardiovascular disease and diabetes mellitus. Gene and chromosome mapping are tools used to develop detection, monitoring, diagnosis and treatment regimens for persons suffering from genetic diseases. The advances in healthcare owe much of their success to the ability of geneticists to view genomes of organisms and analyze chromosomes at a level that allows insight into the transmission and manifestation of genetic diseases.
Before we go further, let us review some basics about chromosomes and genes. A functional chromosome requires four features, as shown in Figure 12.1.1.
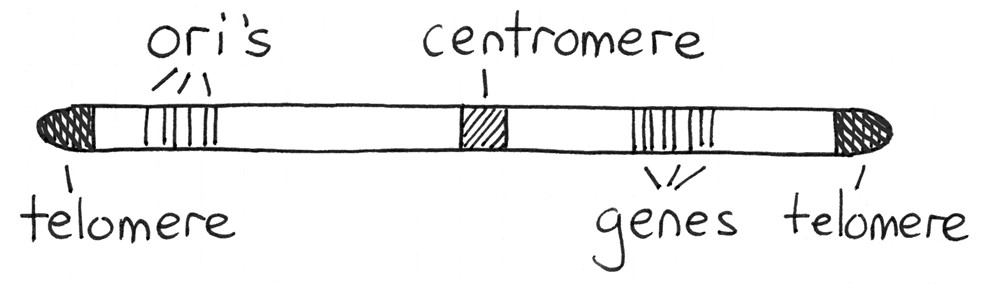
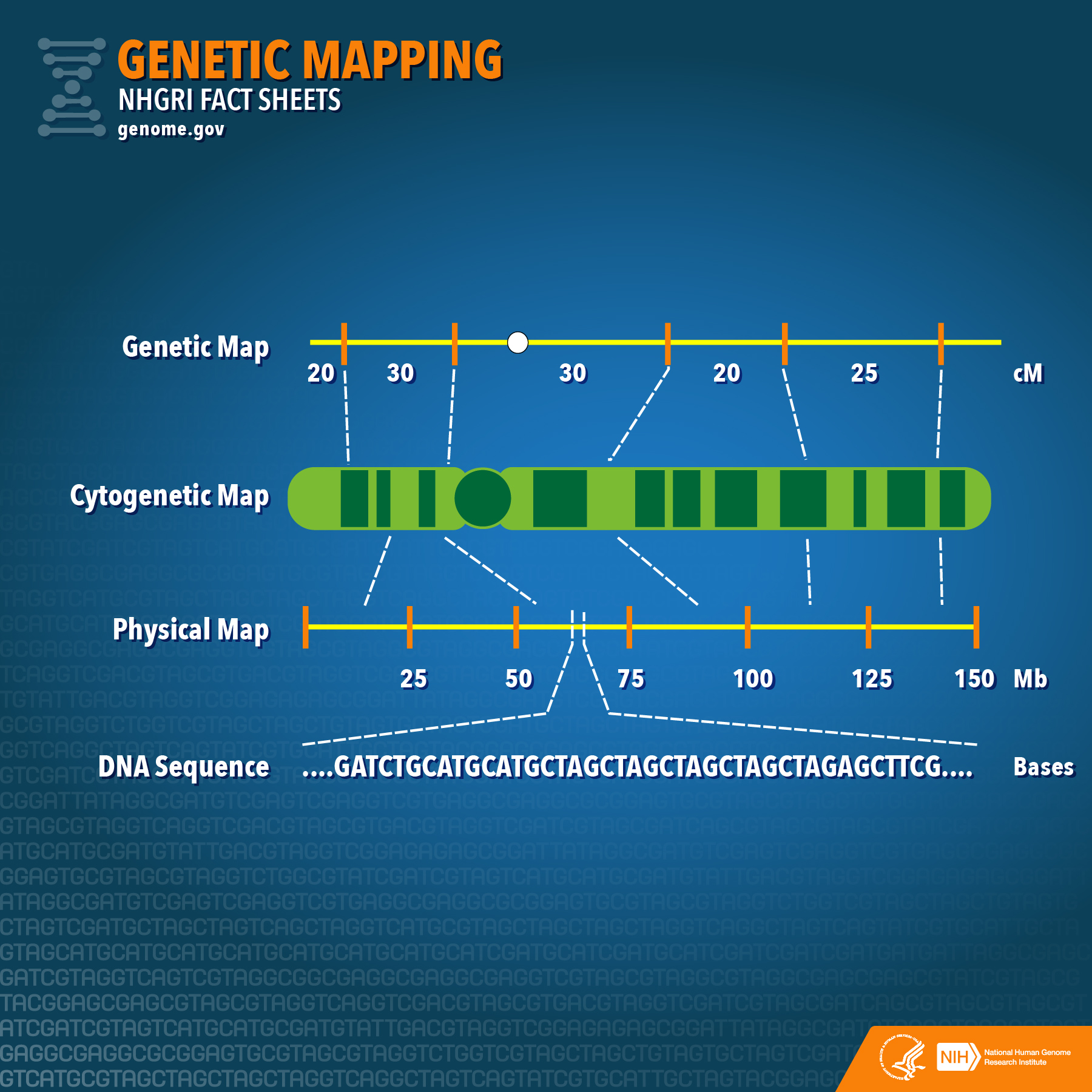
Each chromosome is a long molecule of double-stranded DNA. They carry genetic information (genes). Chromosome 1, our largest chromosome, has the most genes — about 4778 in total. Many of these genes are transcribed into mRNAs, which encode proteins. Other genes are transcribed into tRNAs, rRNA, and other non-coding RNA molecules. A centromere (“middle part”) is a place where proteins attach to the chromosome as required during the cell cycle. Cohesin proteins hold the sister chromatids together beginning in the S phase. Kinetochore proteins form attachment points for microtubules during mitosis. All human chromosomes have a centromere, but not necessarily in the middle of the chromosome. If it is in the centre of the chromosome, it is called a metacentric chromosome. If it is offset a bit, it is submetacentric, and if it is towards one end, the chromosome is acrocentric. In humans, an example of each is chromosome 1, 5, and 21, respectively. Humans do not have any telocentric chromosomes, those with the centromere at one end, but mice and some other mammals do. The ends of a chromosome are called telomeres (“end parts”). Part of the DNA replication is unusual here, it is done with a dedicated DNA polymerase known as a Telomerase. As with the centromere region, there are no genes in the telomeres, just simple, repeated DNA sequences. At the beginning of S phase, DNA polymerases begin the process of chromosome replication. The sites where this begins are called origins of replication (ori’s). They are found distributed along the chromosome, about 40 kb apart. The S phase begins at each ori as two replication forks leave, travelling in opposite directions. Replication continues, and replication forks travelling from one ori will collide with forks travelling towards it from the neighboring ori. When all the forks meet, DNA replication will be complete. Chromosomes are long duplex molecules of DNA that are either linear or circular, and composed of a relatively constant sequence of nucleotides. There are three ways of describing the linear contents of a chromosome (Figure 12.1.2): (1) genetic map, (2) cytogenetic map, and (3) physical map (ultimately the sequence).
Media Attributions
- Figure 12.1.1 Original by M. Harrington (2017), CC BY-NC 3.0, Open Genetics Lectures
- Figure 12.1.2 NHGRI Fact Sheet- Genetic Mapping (27058469495) by National Human Genome Research Institute (NHGRI) Image Gallery, CC BY 2.0, via Wikimedia Commons
Reference
Harrington, M. (2017). Figure 6. Parts of a typical human nuclear chromosome …[digital image]. In Locke, J., Harrington, M., Canham, L. and Min Ku Kang (Eds.), Open Genetics Lectures, Fall 2017 (Chapter 15, p. 4). Dataverse/ BCcampus. http://solr.bccampus.ca:8001/bcc/file/7a7b00f9-fb56-4c49-81a9-cfa3ad80e6d8/1/OpenGeneticsLectures_Fall2017.pdf
Long Descriptions
- Figure 12.1.1 Parts of a typical human nuclear chromosome are shown. The origins of replication and genes are distributed everywhere along the chromosome, except for the telomeres and centromere. Telomeres occur at the ends, and the centromere is shown near the middle of this chromosome. [Back to Figure 12.1.1]
- Figure 12.1.2 A comparison of genetic maps, which show map distances in centimorgans, to cytogenetic maps, which show bands against contrasting colour which represent genes along the chromosome, to physical maps, which show map distances in megabase pairs, to finally the actual DNA sequence of the chromosome itself. Genetic mapping offers firm evidence that a disease transmitted from parent to child is linked to one or more genes. Mapping provides clues about which chromosome contains the gene, and precisely where the gene lies on that chromosome. Genetic maps have been used successfully to find the gene responsible for relatively rare, single-gene inherited disorders, such as cystic fibrosis and Duchenne muscular dystrophy. [Back to Figure 12.1.2]

