Chapter 7 Answers
-
-
-
- Mutant strain #1 has a mutation in gene B (but genes A and C should be functional).
- Mutant strain #2 is in gene C (but genes A and B should be functional).
- Mutant strain #3 is in gene A (but genes B and C should be functional).
- Even prototrophs cannot produce the vitamin biotin, so it must be added for any strain to grow. Wild type strains also lack the enzyme(s) for this biochemical pathway. Biotin is present in Complete Medium.
- No, we now know that genes also encode tRNA, rRNA, and a variety of other functional RNAs.
-
- Changes in many amino acids do not cause a change in function. A specific amino acid is not required at that site for function to occur.
- Changes in many amino acids can cause a minor loss in function. A specific amino acid at a site may be required for optimal function to occur.
- Changes in some amino acids can cause a complete loss in function. Many specific amino acid are required at specific sites for any function to occur (e.g., the active site within an enzyme).
- Any one of the amino acids changed in part (c) can result in a complete loss of function.
- No, the gene can be transcribed into an mRNA and translated into a polypeptide, but the polypeptide is not functional because of a change in an amino acid.
- Chain A has ~268, while chain B has 450. The entire enzyme has ~ 4 chains, two A and two B (a heterodimer).
-
- row 1 orange, orange, orange
- row 2 white, orange, orange
- row 3 yellow, yellow, orange
- row 4 white, yellow, orange
-
- One possible explanation is that original mutagenesis resulted in a loss-of-function mutation in a gene that is essential for early embryonic development, and that this mutation is X-linked recessive in the female. Because half of the sons will inherit the X chromosome that bears this mutation, half of the sons will fail to develop beyond very early development and will not be detected among the F1 The proportion of male flies that were affected depends on what fraction of the female parent’s gametes carried the mutation. In this case, it appears that half of the female’s gametes carried the mutation.
- To test whether a gene is X-linked, you can usually do a reciprocal cross. However, in this case it would be impossible to obtain adult male flies that carry the mutation; they are dead. If the hypothesis proposed in a) above is correct, then half of the females, and none of the living males in the F1 should carry the mutant allele. You could therefore cross F1 females to wild type males, and see whether the expected ratios were observed among the offspring (e.g., half of the F1 females should have a fewer male offspring than expected, while the other half of the F1 females and all of the males should have a roughly equal numbers of male and female offspring).
-
- Treat a population of seeds with a mutagen such as EMS. Allow these seeds to self-pollinate, and then allow the F1 generation to also self-pollinate. In the F2 generation, smell each flower to find individuals with abnormal scent.
- The fishy gene appears to be required to make the normal floral scent. Because the flowers smell fishy in the absence of this gene, one possibility explanation of this is that fishy makes an enzyme that converts a fishy-smelling intermediate into a chemical that gives flowers their normal, sweet smell.
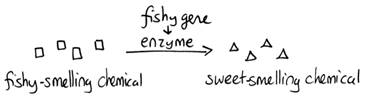 Note that although we show this biochemical pathway as leading from the fishy-smelling chemical to the sweet-smelling chemical in one step, it is likely that there are many other enzymes that act after the fishy enzyme to make the final, sweet-smelling product. In either case, blocking the pathway at the step catalyzed by the fishy enzyme would explain the fishy smell.
Note that although we show this biochemical pathway as leading from the fishy-smelling chemical to the sweet-smelling chemical in one step, it is likely that there are many other enzymes that act after the fishy enzyme to make the final, sweet-smelling product. In either case, blocking the pathway at the step catalyzed by the fishy enzyme would explain the fishy smell. - In nosmell plants, the normal sweet smell disappears. Unlike fishy, the sweet smell is not replaced by any intermediate chemical that we can easily detect. Thus, we cannot conclude where in the biochemical pathway the nosmell mutant is blocked; nosmell may normally therefore act either before or after fishy normally acts in the pathway:
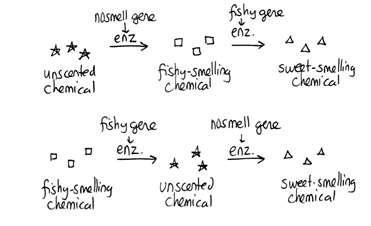 Alternatively, nosmell may not be part of the biosynthetic pathway for the sweet smelling chemical at all. It is possible that the normal function of this gene is to transport the sweet-smelling chemical into the cells from which it is released into the air, or maybe it is required for the development of those cells in the first place. It could even be something as general as keeping the plants healthy enough that they have enough energy to do things like produce floral scent.
Alternatively, nosmell may not be part of the biosynthetic pathway for the sweet smelling chemical at all. It is possible that the normal function of this gene is to transport the sweet-smelling chemical into the cells from which it is released into the air, or maybe it is required for the development of those cells in the first place. It could even be something as general as keeping the plants healthy enough that they have enough energy to do things like produce floral scent.
-
- Dominant mutations are generally much rarer than recessive mutations. This is because mutation of a gene tends to cause a loss of the normal function of this gene. In most cases, having just one normal (wt) allele is sufficient for normal biological function, so the mutant allele is recessive to the wt allele. Very rarely, rather than destroying normal gene function, the random act of mutation will cause a gene to gain a new function (e.g., to catalyze a new enzymatic reaction), which can be dominant (since it performs this new function whether the wt allele is present or not). This type of gain-of-function dominant mutation is very rare because there are many more ways to randomly destroy something than by random action to give it a new function (think of the example given in class of stomping on an iPod).
- Dominant mutations should be detectable in the F1 generation, so the F1 generation, rather than the F2 generation can be screened for the phenotype of interest.
- Large deletions, such as those caused by some types of radiation, are generally less likely than point mutations to introduce a new function into a protein: it is hard for a protein to gain a new function if the entire gene has been removed from the genome by deletion.
-
- Mutagenize a wild type (auxotrophic) strain and screen for mutations that fail to grow on minimal media, but grow well on minimal media supplemented with proline.
- Take mutants #1-#10) and characterize them, based on:
- genetic mapping of the mutants (different locations indicate different genes);
- different response to proline precursors (a different response suggests different genes);
- complementation tests among the mutations (if they complement then they are mutations in different genes).
- If the mutations are in different genes then the F1 progeny would be wild type (able to grow on minimal medium without proline).
- If the mutations are in the same gene then the F1 progeny would NOT be wild type (unable to grow on minimal medium without proline).
-
-

