5.4 Transformation Rescue
Beadle and Tatum built on this connection between genes and metabolic pathways. Their research led to the “one gene, one enzyme (or protein)” hypothesis, which states that each enzyme that acts in a biochemical pathway is encoded by a different gene. Although we now know of many exceptions to the “one gene, one enzyme” principle, it is generally true that each different gene produces a protein with a distinct catalytic, regulatory, or structural function.
Beadle and Tatum used the fungus Neurospora crassa (a bread mould) for their studies, because it had practical advantages as a laboratory model organism. They knew that Neurospora was prototrophic, meaning it could grow on minimal medium (MM). Minimal medium lacked most nutrients, except for a few minerals, simple sugars, and one vitamin (biotin).
Prototrophs can synthesize the amino acids, vitamins, etc., necessary for normal growth.
They also knew that by exposing Neurospora spores to X-rays, they could randomly induce mutations in genes (now known as damage to the DNA leading to DNA sequence change). Each spore exposed to X-rays potentially contained a mutation in a different gene. While most mutagenized spores were still able to grow (prototrophic), some spores had mutations that changed their phenotype from a prototroph into an auxotrophic strain, which could no longer grow on minimal medium. Instead, these auxotrophs could grow on complete medium (CM), which was MM supplemented with nutrients, such as amino acids and vitamins, etc. (Figure 5.4.1). In fact, some auxotrophic mutations could grow on minimal medium with only one single nutrient supplied, such as the amino acid arginine. This implied that each auxotrophic mutant was blocked at a specific step in a biochemical pathway, and that by adding an essential compound, such as arginine, that block could be circumvented.
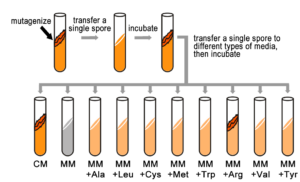
Beadle and Tatum’s experiments are important not only for their conceptual advances in understanding genes, but also because they demonstrate the utility of screening for genetic mutants to investigate a biological process – genetic analysis. Beadle and Tatum’s results were useful to investigate biological processes, specifically the metabolic pathways that produce amino acids. For example, Srb and Horowitz, in 1944, tested the ability of the amino acids to rescue auxotrophic strains. They added one of each of the amino acids to minimal medium and recorded which of these restored growth to independent mutants.
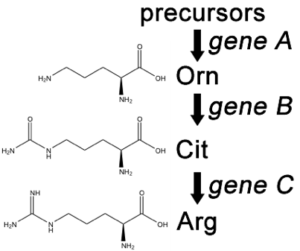
A convenient example is arginine. If the progeny of a mutagenized spore could grow on minimal medium only when it was supplemented with arginine (Arg), then the auxotroph must bear a mutation in the Arg biosynthetic pathway and was called an “arginineless” strain (arg-). Synthesis of even a relatively simple molecule, such as arginine, requires many steps, each with a different enzyme. Each enzyme works sequentially on a different intermediate in the pathway (Figure 5.4.2). For arginine (Arg), two biochemical intermediates are ornithine (Orn) and citrulline (Cit). Thus, mutation of any one of the enzymes in this pathway could turn Neurospora into an Arg auxotroph (arg-). Srb and Horowitz extended their analysis of Arg auxotrophs by testing the intermediates of amino acid biosynthesis for the ability to restore growth of the mutants (Figure 5.4.3).
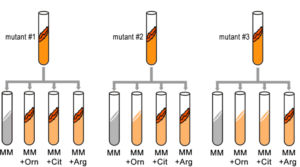
They found that only Arg could rescue all the Arg auxotrophs, while either Arg or Cit could rescue some (Table 5.4.1). Based on these results, they deduced the location of each mutation in the Arg biochemical pathway, (i.e., which gene was responsible for the metabolism of which intermediate).
| Mutants in: | MM + Orn | MM + Cit | MM + Arg |
|---|---|---|---|
| Gene A | Yes | Yes | Yes |
| Gene B | No | Yes | Yes |
| Gene C | No | No | Yes |
In a normal rescue experiment, arginine auxotrophic strands of single-celled Neurospora crassa were “rescued” when supplemented with the amino acids that they could not synthesize and that were essential for the organism’s metabolism. In transformation rescue, rather than giving supplementary metabolic pathway products, it supplies the needed genes that can complement the mutant allele. The process of taking in foreign DNA (transformation) that contains the normal version of the gene and thereby rescuing the auxotrophic strain is called transformation rescue.
Let’s say that there is an E. coli auxotrophic mutant in a gene called “a” (Table 5.4.2).
| E. coli Strain | MM (Minimal medium) | MM + supplement |
|---|---|---|
| a- | Auxotrophic (no growth) | Growth |
| a+ | Growth | Growth |
In order to transform this auxotrophic strain and rescue, we need to:
- Make the E. coli auxotrophic cells competent so that it can incorporate foreign DNA molecules. We can form a competent cell via heat shock or electroporation that can slightly damage the membrane and therefore provide passageways for DNA molecules to enter the cell.
- Extract DNA molecules from a wild type strain of E. coli and break them down into short fragments using enzymes.
- Insert these short fragments of E. coli DNA into a DNA vector, which is a DNA molecule that can contain, amplify, and transfer the inserted DNA fragments into the host cell. This combined DNA molecule is called recombinant DNA. Plasmids are small circular DNA molecules that are mostly found in bacteria and are suitable as DNA vectors. The [vector + DNA insert] molecule can be replicated, and the result would be multiple clones of the original DNA insert.
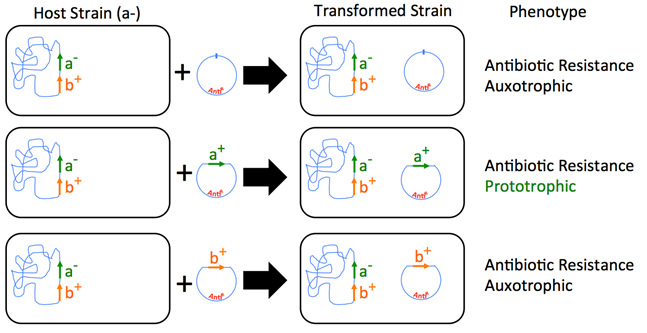
- After the E. coli DNA fragments (that were once a single long DNA molecule) are inserted into DNA vectors, we have a collection of recombinant DNA molecules, which when transformed, can be called a DNA library. Among all the recombinant DNA molecules in the library, there are three possibilities (Figure 5.4.1): (1) DNA clones that contain gene a, (2) DNA clones that don’t contain gene a, which will be collectively presented by the letter b and (3) DNA clones that don’t contain any foreign genes.
- Combine the recombinant DNA molecules and host E. coli strain together, so that the auxotrophic strain can incorporate those DNA molecules through transformation. Growing the strains on minimal and complete media will let us decide if the transformation rescue worked or not.
- The host strain’s genotype is a-b+. It needs a wild type a+ to grow on minimal medium. Therefore, plasmids that have the a+ allele would grow (prototrophic), and other strains that have either a plasmid with no transgene or have a plasmid with gene b+ would be still auxotrophic.
Notice that the plasmids contain an antibiotic resistance gene called AntiR and that the strains were grown on minimal medium that contained antibiotics. Why was this so? This is because we want to select for the ones that incorporated the plasmid that contained the wild-type “a” gene.
Only a small fraction of cells is transformed by foreign DNA. Therefore, if we grow those strains on agar plate without antibiotics, we cannot guarantee the growth was due to the complementation between the host DNA and the recombinant DNA or by some reversion back to wild type. There is a small possibility that the cells that weren’t transformed could somehow synthesize the essential substrate due to a spontaneous mutation. Adding the antibiotic selection will remove cells that weren’t transformed and, therefore, don’t contain a plasmid with the antibiotic resistance gene, and select for the cells that were successfully transformed and complemented by the recombinant DNA.
Media Attributions
- Figure 5.4.1 Original by Deyholos (2017), CC BY-NC 3.0, Open Genetics Lectures
- Figure 5.4.2 Original by Deyholos (2017), CC BY-NC 3.0, Open Genetics Lectures
- Figure 5.4.3 Original by Deyholos (2017), CC BY-NC 3.0, Open Genetics Lectures
- Figure 5.4.4 Original by Locke (2017), CC BY-NC 3.0, Open Genetics Lectures
References
Deyholos, M. (2017). Figures: 3. A single mutagenized spore; 4. A simplified version of the Arg biosynthetic pathway…; and 5. Testing different Arg auxotrophs… [image]. In Locke, J., Harrington, M., Canham, L. and Min Ku Kang (Eds.), Open Genetics Lectures, Fall 2017 (Chapter 3, p. 3). Dataverse/ BCcampus. http://solr.bccampus.ca:8001/bcc/file/7a7b00f9-fb56-4c49-81a9-cfa3ad80e6d8/1/OpenGeneticsLectures_Fall2017.pdf
Locke, J. (2017). Figure 7. Transformation rescue diagram [digital image]. In Locke, J., Harrington, M., Canham, L. and Min Ku Kang (Eds.), Open Genetics Lectures, Fall 2017 (Chapter 4, p. 5). Dataverse/ BCcampus. http://solr.bccampus.ca:8001/bcc/file/7a7b00f9-fb56-4c49-81a9-cfa3ad80e6d8/1/OpenGeneticsLectures_Fall2017.pdf
Long Descriptions
- Figure 5.4.1 Production of a colony of genetically identical fungi. First, in a test tube, the sample is mutagenized, and then a single spore is transferred to another test tube , which is then incubated. Then, we transfer a single spore to different types of media, and incubate. Spores are Tested for Their Ability to Grow on Different Types of Media. Because spores of this particular colony are able to grow only on complete medium (CM), or on minimal medium supplemented with arginine (MM+Arg), they are considered Arg auxotrophs and we infer that they have a mutation in a gene in the Arg biosynthetic pathway. This type of screen is repeated many times to identify other mutants in the Arg pathway and in other pathways. [Back to Figure 5.4.1]
- Figure 5.4.2 Three organic structures – ornithine, citrulline and Arginine. From a precursor, Gene A is able to facilitate synthesis of ornithine. Gene B then converts it into Citrulline. Finally, gene C converts this into Arginine. This biosynthetic pathway shows the involvement of multiple genes in the synthesis of nutrients within a cell. [Back to Figure 5.4.2]
- Figure 5.4.3 Three test tubes labelled mutant 1, 2 and 3 respectively. In each case, the mutants are subjected to four different types of media – that is, each is supplemented with a different combination of nutrients. The media are minimal media (MM), MM + ornithine, MM + citrulline and MM + arginine. The ability (or not) of the mutants to proliferate within the various media is indictive of their genotypes. [Back to Figure 5.4.3]
- Figure 5.4.4 The differences between auxotrophic and prototrophic organisms. A plasmid carrying an a+ gene is used to rescue an a- strain. The product of the a+ strain is an enzyme, which then confers antibiotic resistance. Adding the antibiotic selection will remove cells that weren’t transformed and, therefore, don’t contain a plasmid with the antibiotic resistance gene, and select for the cells that were successfully transformed and complemented by the recombinant DNA. [Back to Figure 5.4.4]

