Chapter 7 Study Questions
- Compare Figure 7.3.1 and Table 7.3.1 in Section 7.3 Suppose you created three new arg– mutation called mutants #1, #2, & #3. #1 grew on MM+cit and MM+arg, #2 grew on only MM+arg, while #3 grew on MM+ orn, cit or arg. Which genes are #1, 2, & 3 mutant in (A, B, or C)?
- Why was the Vitamin biotin always added the MM?
- Last century, A. Garrod, and later Beadle and Tatum, showed that genes encode enzymes. From what we know now, do all genes encode enzymes? Explain.
- Most mutant proteins differ from wild type (normal) by a single substitution at a specific amino acid site. Explain how some amino acid changes result in:
- no loss of protein function,
- only partial loss-of-function,
- complete loss-of-function,
- and how do changes at different amino acid sites result in the same complete loss-of-function.
- Some mutants result in the loss of a specific enzyme activity. Does this mean that no protein product is produced from that mutant gene?
- The molecular weight of the A and B chains of coli tryptophan synthase are 29,500 and 49,500, respectively. The size of the entire enzyme is 158,000.
- If the average molecular weight of each amino acid is 110, then how many amino acids are present in each chain?
- How many chains does the whole enzyme contain? Explain.
- Recall that Neurospora is orange coloured bread mould. This biochemical pathway below is how wild type cells become orange. None of the compounds are essential. Cells containing W are white, cells with Y are yellow, and cells with O are orange. Assume that the reactions will go to completion, if possible.
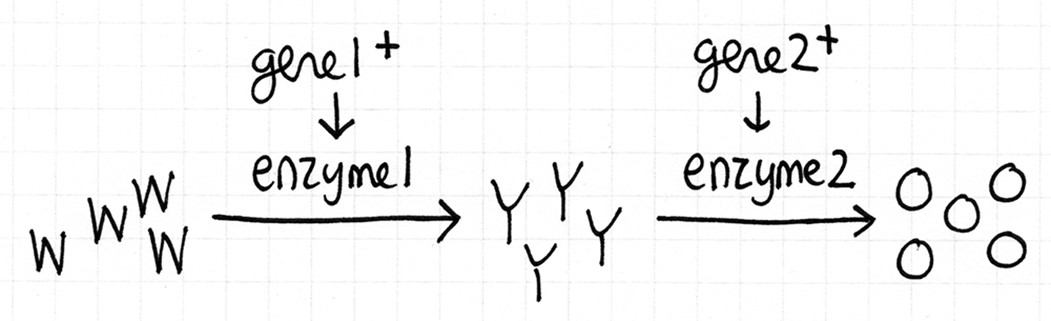 Fill in this table with the colours of the cell cultures.
Fill in this table with the colours of the cell cultures.
Strain MM+W MM+Y MM+O gene1+ gene2+
gene1- gene2+
gene1+ gene2-
gene1- gene2-
- You have a female fruit fly, whose father was exposed to a mutagen (she, herself, wasn’t). Mating this female fly with another non-mutagenized, wild-type male produces offspring that all appear completely normal, except there are twice as many daughters as sons in the F1 progeny of this cross.
- Propose a hypothesis to explain these observations.
- How could you test your hypothesis?
- You decide to use genetics to investigate how your favourite plant makes its flowers smell good.
- What steps will you take to identify genes required for production of the sweet floral scent? Assume this plant is a self-pollinating diploid.
- One of the recessive mutants you identified has fishy-smelling flowers, so you name the mutant (and the mutated gene) fishy. What do you hypothesize about the normal function of the wild-type fishy gene?
- Another recessive mutant lacks floral scent altogether, so you call it nosmell. What could you hypothesize about the normal function of this gene?
- Suppose you are only interested in finding dominant mutations that affect floral scent.
- What do you expect to be the relative frequency of dominant mutations, compared to recessive mutations, and why?
- How will you design your screen differently than in the previous question, to detect dominant mutations specifically?
- Which kind of mutagen is most likely to produce dominant mutations, a mutagen that produces point mutations, or a mutagen that produces large deletions?
- You are interested in finding genes involved in synthesis of proline (Pro), an amino acid normally synthesized by a particular model organism.
- How would you design a mutant screen to identify genes required for Pro synthesis?
- Imagine your screen identified ten mutants (#1 through #10) that grew poorly unless supplemented with Pro. How could you determine the number of different genes represented by these mutants?
- If each of the four mutants represents a different gene, what will be the phenotype of the F1 progeny if any pair of the four mutants are crossed?
- If each of the four mutants represents the same gene, what will be the phenotype of the F1 progeny if any pair of the four mutants are crossed?

